Description & Citation
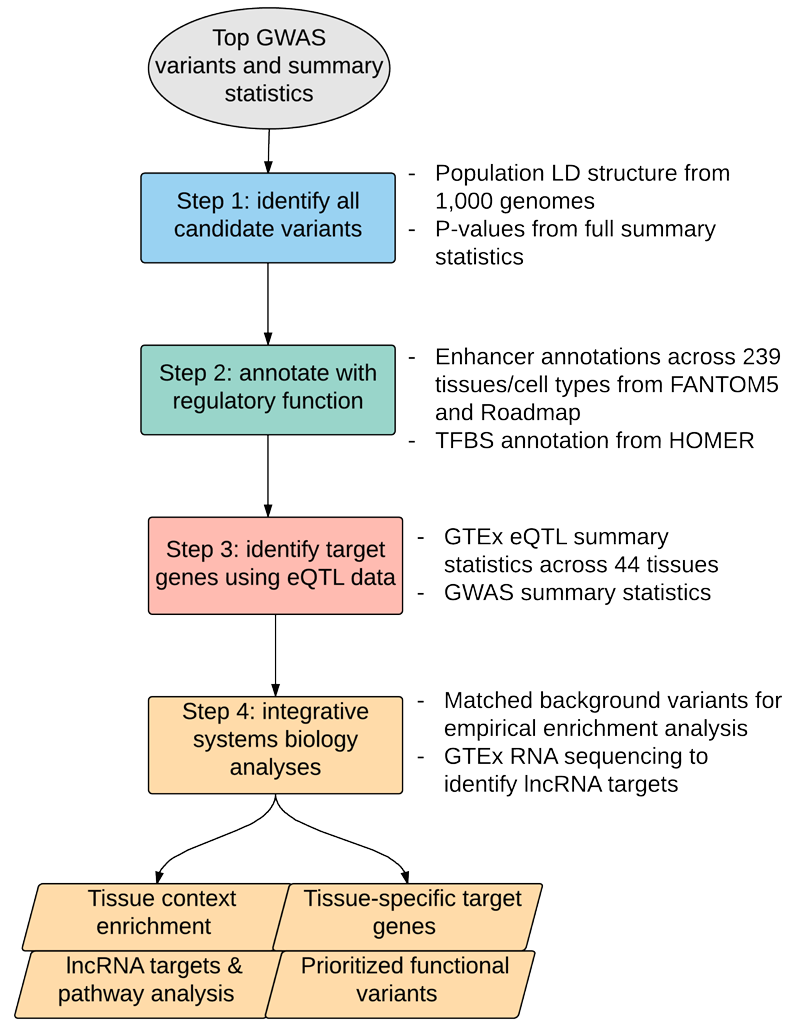
The majority of variants identified by genome-wide association studies (GWAS) reside in the noncoding genome, where they affect regulatory elements including transcriptional enhancers. We propose INFERNO (INFERring the molecular mechanisms of NOncoding genetic variants), a novel method which integrates hundreds of diverse functional genomics data sources with GWAS summary statistics to identify putatively causal noncoding variants underlying association signals. INFERNO comprehensively characterizes the relevant tissue contexts, target genes, and downstream biological processes affected by functional variants.
- Sets of all putatively causal variants are generated by p-value and LD expansion
- All variants in expanded sets are overlapped with functional genomics annotations spanning 239 tissues and cell types from FANTOM5 and Roadmap, as well as with transcription factor binding sites identified by HOMER
- Tissues and cell types from each functional genomics data source are grouped into 32 broad tissue categories for integrative analysis across diverse functional genomics data sources
- Empirical p-values for the enrichment of functional overlaps in each tissue category are obtained by sampling control variants matched on LD block size, distance to nearest gene, and MAF
- To improve on direct eQTL overlap, which is biased by LD structure, INFERNO applies the COLOC Bayesian method for co-localization analysis of GWAS and GTEx eQTL signals
- Co-localization analysis often identifies lncRNA eQTL targets, so GTEx RNA-seq data across 11,439 tissue samples is used to compute lncRNA - mRNA expression correlations and identify targeted mRNAs
INFERNO method paper is now published in Nucleic Acids Research: https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gky686/5064786.
Alexandre Amlie-Wolf, Mitchell Tang, Elisabeth E Mlynarski, Pavel P Kuksa, Otto Valladares, Zivadin Katanic, Debby Tsuang, Christopher D Brown, Gerard D Schellenberg, Li-San Wang. INFERNO: inferring the molecular mechanisms of noncoding genetic variants. Nucleic Acids Research, 2018, gky686, https://doi.org/10.1093/nar/gky686
Note that items 4-7 are only provided in the full pipeline as they are too computationally intensive to run on this web server.
Detailed description of the full INFERNO pipeline is available in Using INFERNO to Infer the Molecular Mechanisms Underlying Noncoding Genetic Associations paper:
Amlie-Wolf, A., Kuksa, P.P., Lee, C.-Y., Mlynarski, E., Leung, YY, Wang, L.-S. Using INFERNO to Infer the Molecular Mechanisms Underlying Noncoding Genetic Associations. Methods Mol. Biol. 2254, 73–91 (2021). PMID: 33326071
Kuksa, P. P., Lee, C.-Y., Amlie-Wolf, A., Gangadharan, P., Mlynarski, E. E., Chou, Y.-F., Lin, H.-J., Issen, H., Greenfest-Allen, E., Valladares, O., Leung, YY, Wang, L.-S. SparkINFERNO: A scalable high-throughput pipeline for inferring molecular mechanisms of non-coding genetic variants. Bioinformatics (2020) doi:10.1093/bioinformatics/btaa246. PMID: 32330239